mkdir -p course_data/Anterior
mkdir -p course_data/Posterior
# Anterior
cd course_data/Anterior
LINK_PREFIX=https://cf.10xgenomics.com/samples/spatial-exp/1.1.0/V1_Mouse_Brain_Sagittal_Anterior/V1_Mouse_Brain_Sagittal_Anterior
curl -o filtered_feature_bc_matrix.h5 "$LINK_PREFIX"_filtered_feature_bc_matrix.h5
curl -o spatial.tar.gz "$LINK_PREFIX"_spatial.tar.gz
curl -o web_summary.html "$LINK_PREFIX"_web_summary.html
tar -xf spatial.tar.gz
rm spatial.tar.gz
# Posterior
cd ../Posterior
LINK_PREFIX=https://cf.10xgenomics.com/samples/spatial-exp/1.1.0/V1_Mouse_Brain_Sagittal_Posterior/V1_Mouse_Brain_Sagittal_Posterior
curl -o filtered_feature_bc_matrix.h5 "$LINK_PREFIX"_filtered_feature_bc_matrix.h5
curl -o spatial.tar.gz "$LINK_PREFIX"_spatial.tar.gz
curl -o web_summary.html "$LINK_PREFIX"_web_summary.html
tar -xf spatial.tar.gz
rm spatial.tar.gzSetup
Material
Exercises
Login and set up
Log in to Rstudio server with the provided link, username and password.
Create a project
Now that you have access to an environment with the required installations, we will set up a project in a new directory. On the top right choose the button Project (None) and select New Project…
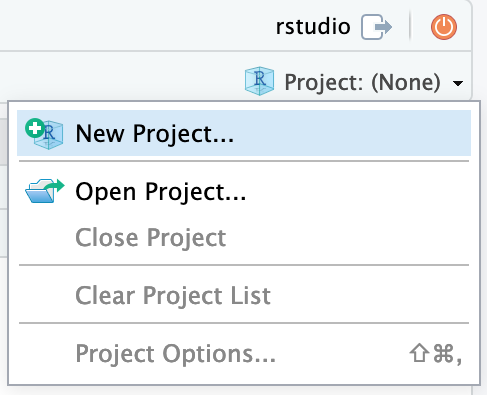
Continue by choosing New Directory
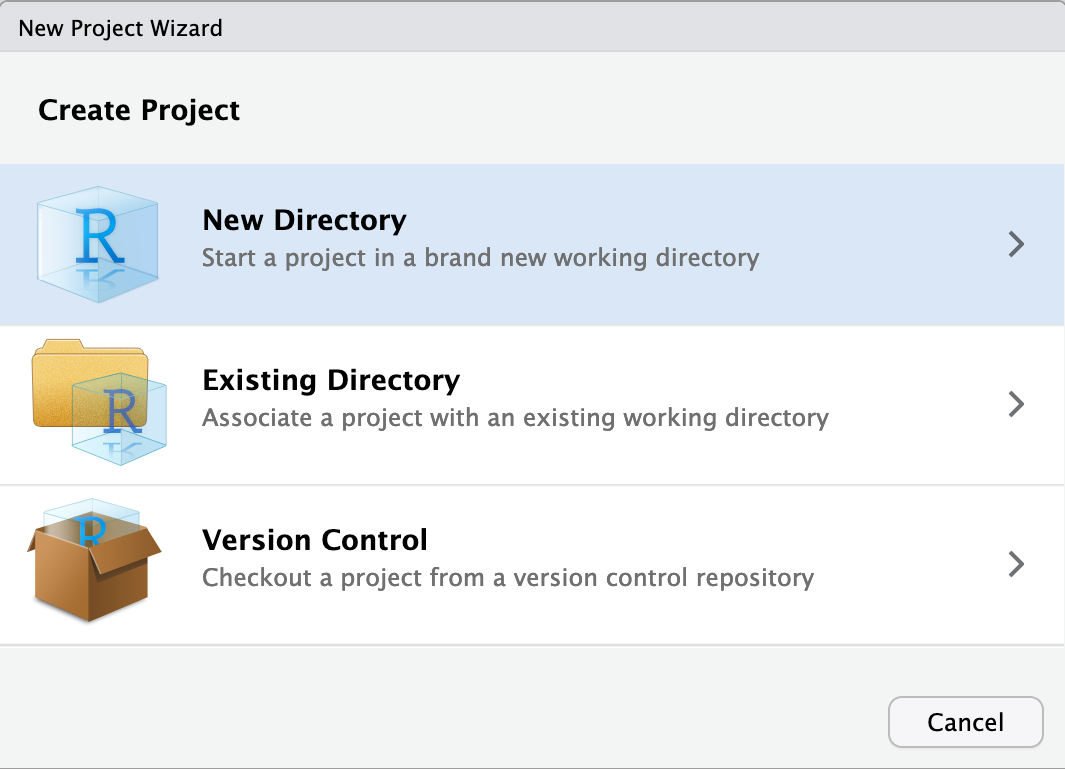
As project type select New Project
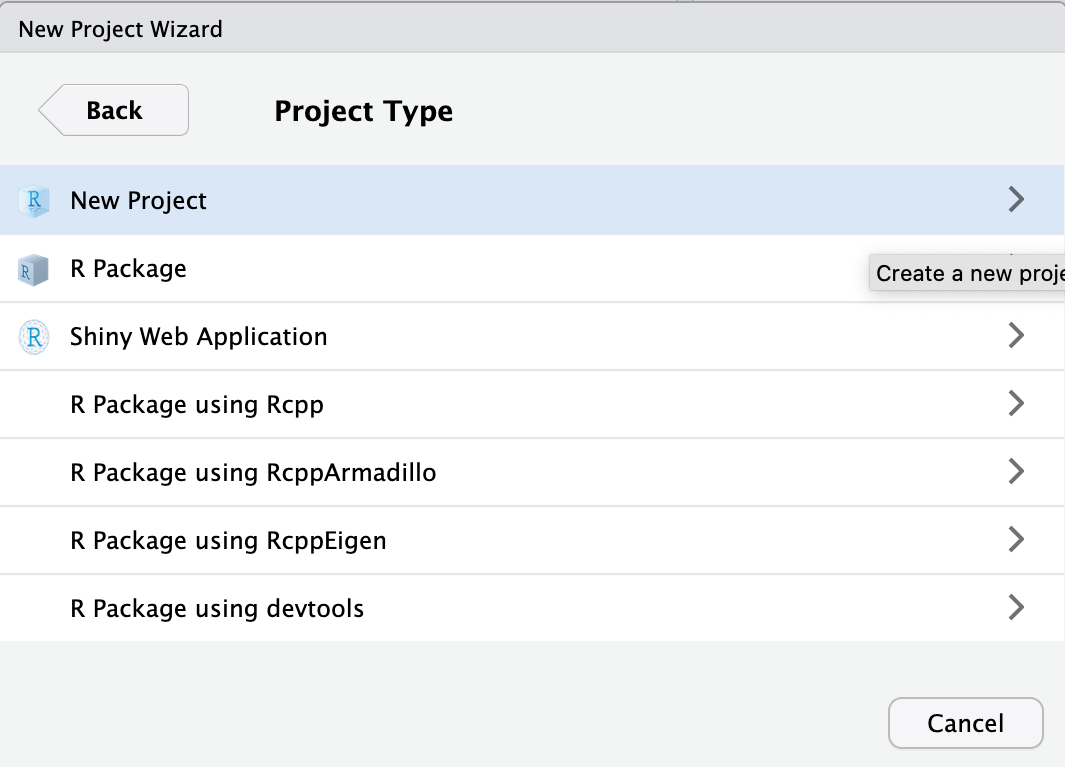
Finally, type in the project name. This should be spatial_transcriptomics_course. Finish by clicking Create Project.
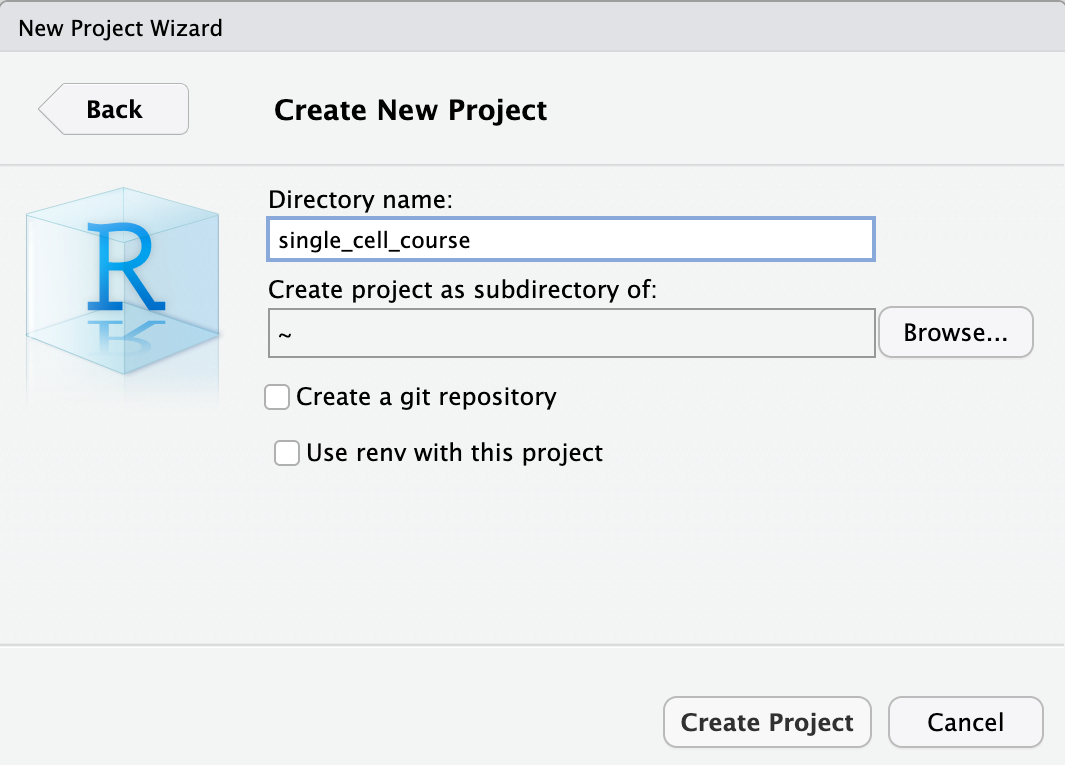
Now that we have setup a project and a project directory (it is in /home/rstudio/spatial_transcriptomics_course), we can download the data that is required for this course. We will use the built-in terminal of Rstudio. To do this, select the Terminal tab:
Downloading the course data
To download and extract the dataset, copy-paste these commands inside the terminal tab:
Have a look at the data directory you have downloaded. It should contain the required data for two slices, the anterior side of a sagittal brain slice, and a posterior side:
course_data/
├── Anterior
│ ├── filtered_feature_bc_matrix.h5
│ ├── spatial
│ │ ├── aligned_fiducials.jpg
│ │ ├── detected_tissue_image.jpg
│ │ ├── scalefactors_json.json
│ │ ├── tissue_hires_image.png
│ │ ├── tissue_lowres_image.png
│ │ └── tissue_positions_list.csv
│ └── web_summary.html
└── Posterior
├── filtered_feature_bc_matrix.h5
├── spatial
│ ├── aligned_fiducials.jpg
│ ├── detected_tissue_image.jpg
│ ├── scalefactors_json.json
│ ├── tissue_hires_image.png
│ ├── tissue_lowres_image.png
│ └── tissue_positions_list.csv
└── web_summary.html
4 directories, 16 filesThis data is provided by 10x genomics, and further information, like material and methods can be found here: