Exercise 7
Enrichment analysis
Load Libraries
OverlapMatrix
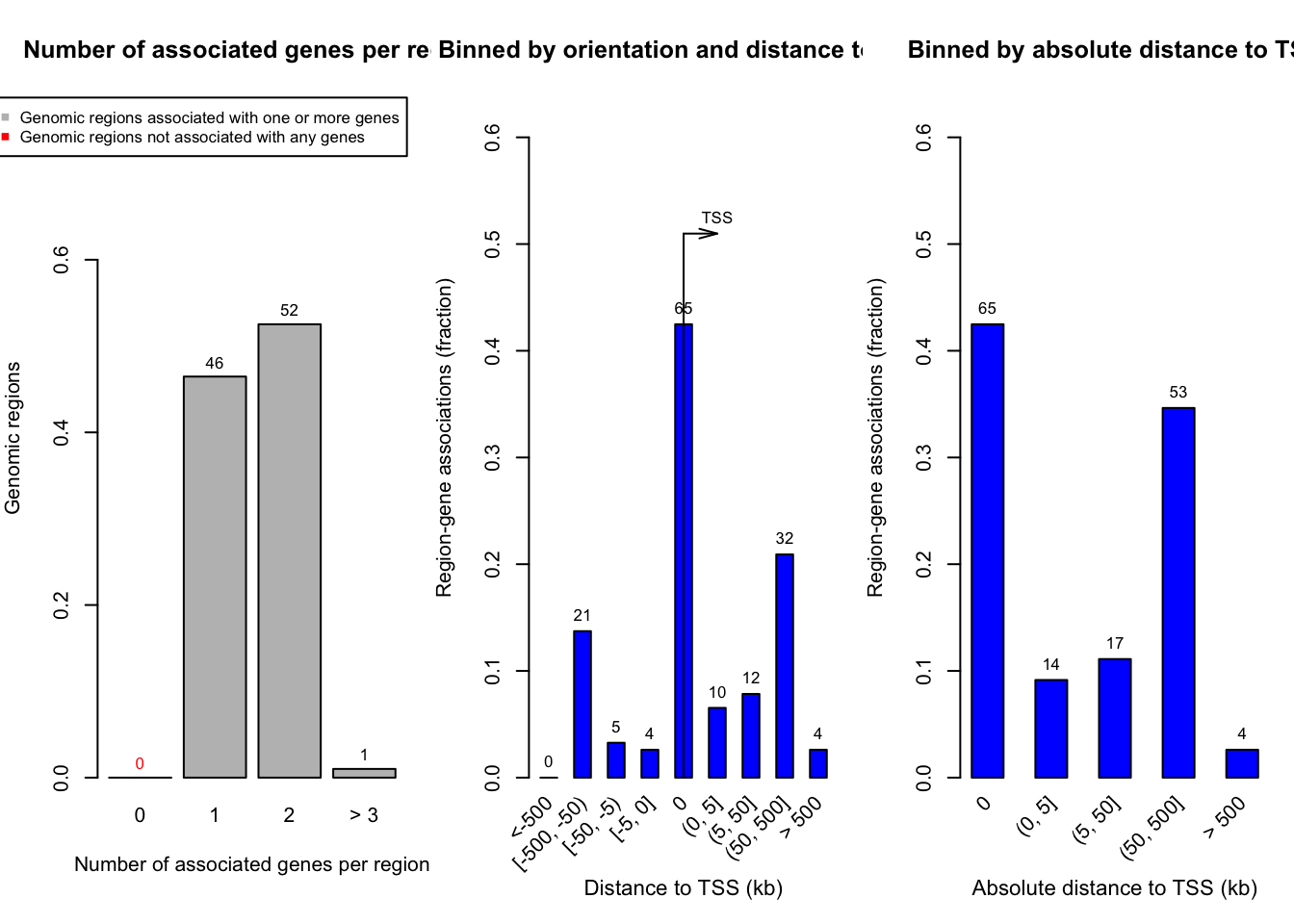
overlap <- readRDS("output/overlap_anno2.rds")We have several cluster of genomic activities. We will run enrichment analysis of regions from one of them, “Active Promoters”.
Running enrichment analysis on regions of our interest
subset the GRanges
gr <- overlap[overlap$ATAC_RNA == "Active Promoters"]
seqlevels(gr) <- as.character(unique(seqnames(gr)))Running enrichment analysis
res <- great(gr = gr, gene_sets = "BP", tss_source = "mm10")* TSS source: TxDb.* check whether TxDb package 'TxDb.Mmusculus.UCSC.mm10.knownGene' is installed.* gene ID type in the extended TSS is 'Entrez Gene ID'.* restrict chromosomes to 'chr1, chr2, chr3, chr4, chr5, chr6, chr7, chr8, chr9, chr10,
chr11, chr12, chr13, chr14, chr15, chr16, chr17, chr18, chr19, chrX, chrY, chrM'.* 20593/24515 protein-coding genes left.* update seqinfo to the selected chromosomes.* TSS extension mode is 'basalPlusExt'.* construct the basal domains by extending 5000bp to upstream and 1000bp to downsteram of TSS.* calculate distances to neighbour regions.* extend to both sides until reaching the neighbour genes or to the maximal extension.* use GO:BP ontology with 15445 gene sets (source: org.Mm.eg.db).* check gene ID type in `gene_sets` and in `extended_tss`.* use whole genome as background.* remove excluded regions from background.* overlap `gr` to background regions (based on midpoint).* in total 99 `gr`.* overlap extended TSS to background regions.* check which genes are in the gene sets.* only take gene sets with size >= 5.* in total 9412 gene sets.* overlap `gr` to every extended TSS.* perform binomial test for each biological term.Results
Volcano plot
plotVolcano(res)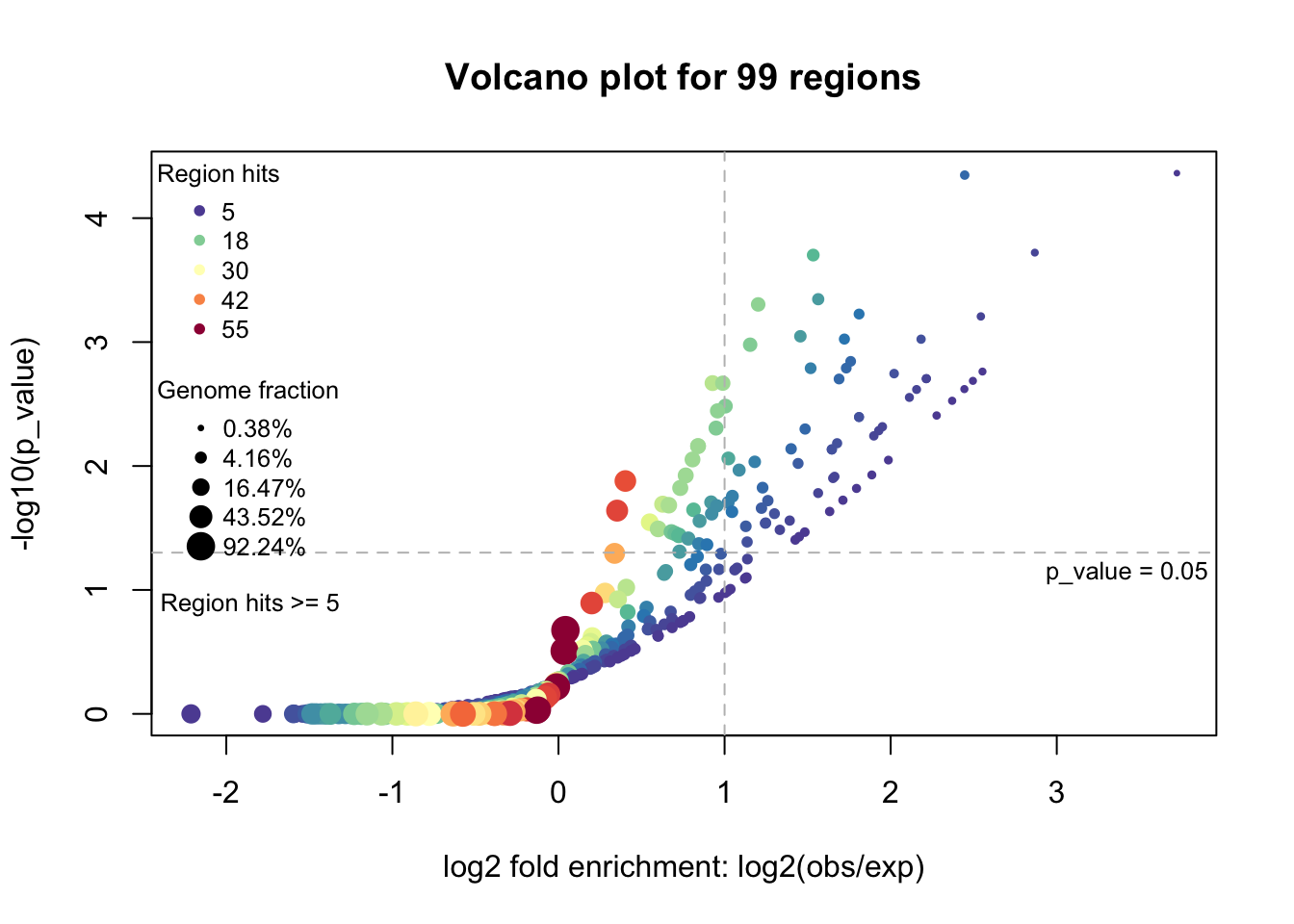
Regions-gene assiciation plot
There is a shiny application to explore the results
shinyReport(res)Simplyfied representation of GO terms
tb <- getEnrichmentTable(res)
sig_go_ids <- tb$id[tb$p_adjust < 0.05]
cl <- simplifyGO(mat = sig_go_ids)You haven't provided value for `ont`, guess it as `BP`.relations: is_a, part_of, regulates, negatively_regulates, positively_regulatesIC_method: IC_annotationterm_sim_method: Sim_XGraSM_2013IC_method: IC_annotationCluster 33 terms by 'binary_cut'... 4 clusters, used 0.1181722 secs.
'magick' package is suggested to install to give better rasterization.
Set `ht_opt$message = FALSE` to turn off this message.
Perform keywords enrichment for 4 GO lists...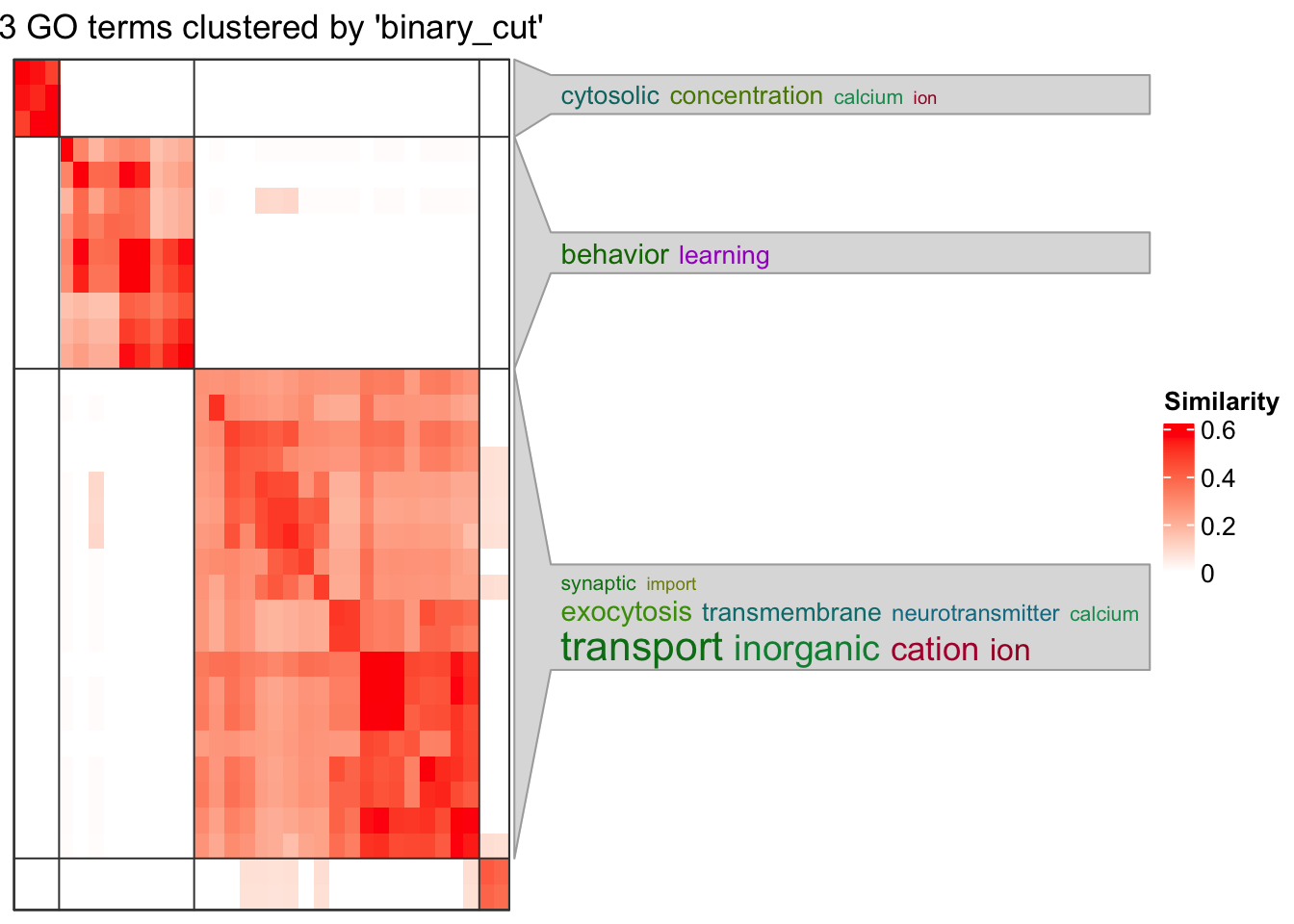
summarizeGO(go_id = sig_go_ids, -log10(tb$p_adjust[tb$p_adjust < 0.05]),
axis_label = "average -log10(p.adjust)")You haven't provided value for `ont`, guess it as `BP`.term_sim_method: Sim_XGraSM_2013IC_method: IC_annotationCluster 33 terms by 'binary_cut'... 4 clusters, used 0.1288469 secs.
Perform keywords enrichment for 4 GO lists...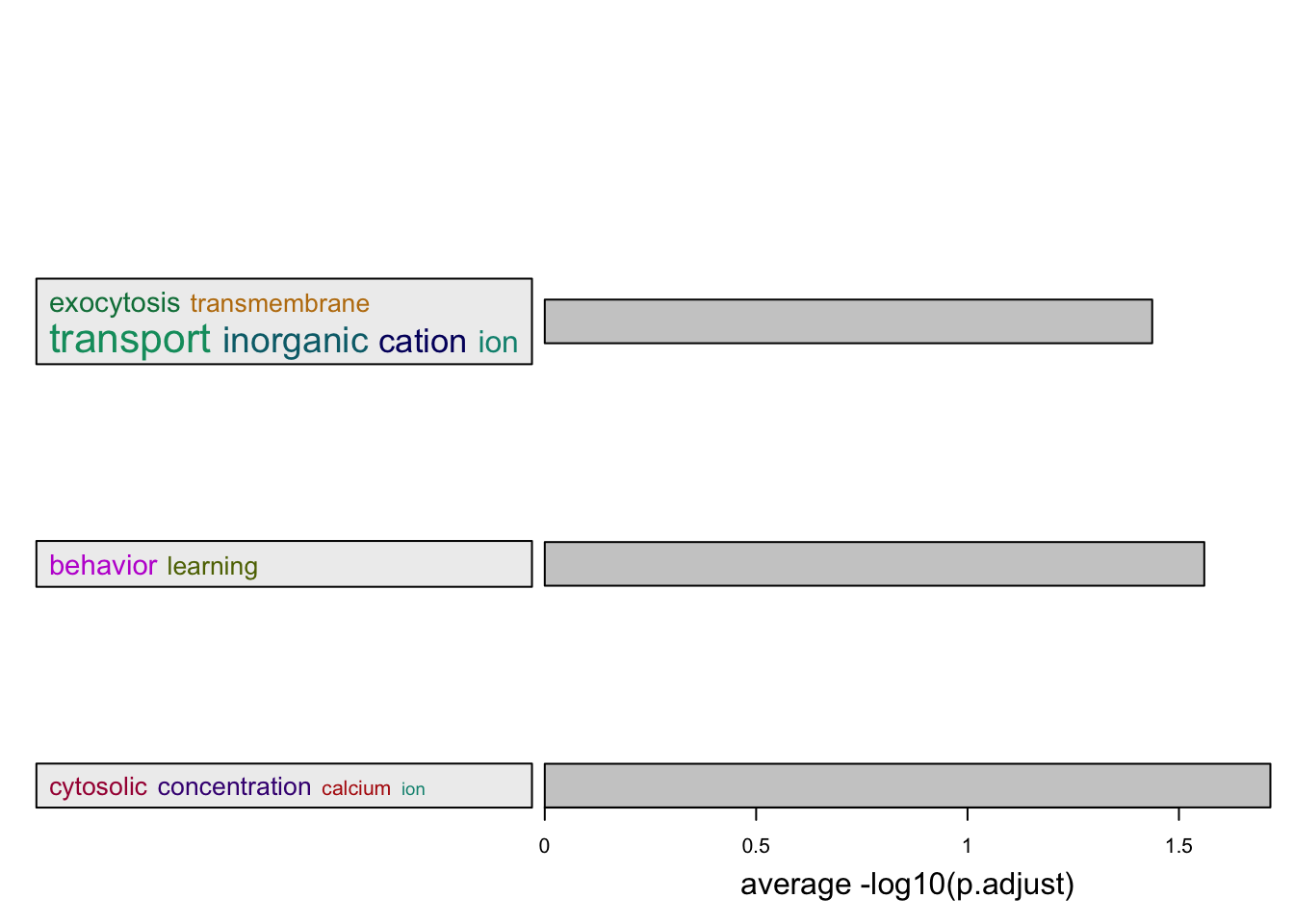
Output
The heatmap shows clear clustering of GO terms into two main clusters, indicating distinct functional groups.
The GO terms on the right highlight biological processes relevant to neuronal and ion transport functions, such as transport, inorganic cation ion, exocytosis, synaptic, neurotransmitter, and behavior learning.
The cluster with terms like cytosolic concentration, calcium ion suggests a calcium-related signaling or ion homeostasis process at play.
The color gradient and similarity scores (up to ~0.6) indicate moderate similarity within clusters, which is reasonable for GO term groupings.
Interpretation
E11.5 to E15.5 is a key window in neural development involving neuronal differentiation, synapse formation, and active signaling.
Active promoters linked to ion transport, synaptic transmission, calcium signaling are expected to be regulated during this developmental stage.
The clustering separating signaling and transport-related terms from behavior and learning terms also aligns with early neural circuit formation and functional maturation.
Question
Perform enrichment analysis of any other category.